Calculating SUVmax for GE’s PET/CT scanner
Abstract
In order to calculate SUVmax for GE data, the following formula has to be utilized:
SUVlbm = [activity, i.e., signal for a single pixel]
* 1.10 * weight – 120 * (weight/height^2)
/
[tracer_activity * 2^( -(scan_time – measured_time)
/
half_life)]To be more precise, below is the same formula, but with more specific information on the voxel and DICOM metadata fields required:
SUVlbm = [PET voxel (raw) data, e.g., from location: x=23, y=62, z=83]
* 1.10 * weight(from:0010,1010)
– 120 * (weight(from:0010,1010)/height(from:0010,1020)^2)
/
[dose(from:0018,1074)
* 2^( -(scan_time(from:0008,0031)
- measured_time(from:0018,1072))
/
half_time(from:0018,1075))]For more information, please refer to the article below.
Introduction
An algorithm for calculating SUVmax value for PET/CT GE scanner will be described in this post.
The starting point for this post is my post at Slicer 3D forum: https://discourse.slicer.org/t/pet-standard-uptake-value-computation/7491/4
Problem description: One would like to know SUV values and SUVmax for a given ROI in PET data from a GE scanner. This of course can be extracted from the GE work station, however, for a more refined analyses, the whole volume with SUVs calculated is required. The GE workstation allows for exporting just the “raw” PET data, i.e., activity counts, not the SUVs (which is a kind of a correction). Activity counts are in a scale of thousands, SUV are in a scale of (maximally) dozens.
Partial solution: Slicer 3D has an algorithm for calculating SUVmax implemented, however: the result are not the same as from the GE work station. See my post: https://discourse.slicer.org/t/pet-standard-uptake-value-computation/7491/4 ; and the implementation of the algorithm: https://github.com/Slicer/Slicer/blob/main/Modules/CLI/PETStandardUptakeValueComputation/PETStandardUptakeValueComputation.cxx#L1454-L1457
Notes on the complete solution
Following this post: https://discourse.slicer.org/t/how-to-calculate-the-suv/16831/2 you have to open the following link: https://qibawiki.rsna.org/index.php/Standardized_Uptake_Value_(SUV) , and download the GE Standard:
The following note assume that you calculate SUV for LBM (lean body mass, lbm(g/ml)* , i.e., for the body mass without the fat).
See the following formula from the GE’s *.doc document:
SUVlbm = pixel * lbm /actual_activity;
lbm = 1.10 * weight – 120 * (weight/height^2) for male;
= 1.07 * weight – 148 * (weight/height^2) for female;
actual activity = [tracer_activity * 2^( -(scan_time – measured_time) / half_life) ]
Let’s focus on the formula for males:
One can reformulate the above expressions as (for male):
SUVlbm = [activity/signal for a single pixel] * 1.10 * weight – 120 * (weight/height^2) / [tracer_activity * 2^( -(scan_time – measured_time) / half_life)]Hopefully, some of the expressions are already implemented in Slicer 3D, and can serve as a validation for our implementation, see, e.g., this line of code from the DecayCorrection function (source: https://github.com/Slicer/Slicer/blob/main/Modules/CLI/PETStandardUptakeValueComputation/PETStandardUptakeValueComputation.cxx#L732) :
double correctedVal = inVal * (double)pow(2.0, -(decayTime / halfLife) );Sidenote: The Slicer’s implementation is also important from the point of view of reading the proper parameters from DICOM’s header.
In other words, in what in GE’s documentation is called actual_activity is called “decay correction” (correctedVal) in Slicer’s implementation.
Additionally, according to Slicer’s implementation, [-(scan_time – measured_time)] == `decayTime`.
How to calculate the “decay time”? Response:
double decayTime = scanTimeSeconds - startTimeSecondsNote: Just be sure to get the `-` (minus) signs right — this has to be validated experimentally/practically.
A brief research into the materials revealed that the difference may be caused by the fact that Slicer’s implementation does simply just:
double weightByDose = weight / dose;
suvmax = (CPETmax * tissueConversionFactor) * weightByDose;While for LBM and GE the formula says:
SUVlbm = pixel * lbm /actual_activity;Ambiguity: what is the difference between the pixel and the tracer_activity in the GE SUV calculation formula? The formula:
SUVlbm = pixel * [1.10 * weight – 120 * (weight/height^2)] / [tracer_activity * 2^( -(scan_time – measured_time) / half_life)]After some analysis of the documentation, I think that the pixel and tracer_activity are the same thing, i.e., the same value. Additionally, to sum up, what is called dose (the “corrected dose”) in Slicer’s implementation, in GE document is: actual_activity (in detail: [tracer_activity * 2^( -(scan_time – measured_time) / half_life)]). Furthermore, Slicer also uses a tissue conversion factor, while GE does not.
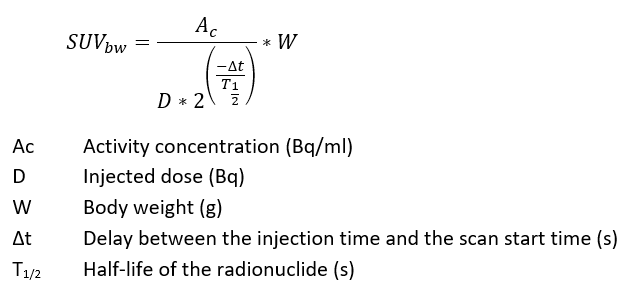
See also this visualization of the formula (for SUVbw):
However, for SUVlbm, instead of the “W” parameter, the “LBM” parameter is used (lean body mass, i.e., without fat).
The important assumption is that what is called CPETmax in Slicer’s implementation is the pixel variable from the GE document. Interestingly, as we said earlier, the most probable interpretation of the GE document formula was that pixel is the same as tracer_activity. This assumption can be validated and tested on real data, using the ideas behind Slicer’s implementation. Meaning, that if pixel and tracer_activity are the same thing (value), then Slicer’s CPETmax and inVal would also be the same. (Optionally, CPETmax * tissueConversionFactor is the same as inVal — less probable, though possible).
Regarding the code in Slicer’s implementation:
labelstat->SetInputConnection(petVolumeConnection);uses petVolumeConnection from line #824
petVolumeConnection = reader1->GetOutputPort();The petVolumeConnection is just the loaded (raw) PET data.
Apart from a very complex logic for masking 3D volume, thresholding, etc., and calculating PETmax (CPETmax), there is an important notion in the Slicer’s code that I noticed earlier, but just now I realised that: in the DecayCorrection function, the inVal is actually the dose. So, in other words, it seems that what is called in the GE document a tracer_activity is actually a dose, i.e., following the Slicer’s code:
dose = ConvertRadioactivityUnits( dose, list.radioactivityUnits.c_str(), "MBq");then:
double dose = list.injectedDose;after:
seq.GetElementDS(0x0018,0x1074,1,&list.injectedDose,false)So, the DICOM metadata location for the dose is (0018, 1074) — 0018,1074 Radionuclide Total Dose: 370500000, see: https://github.com/Slicer/Slicer/blob/main/Modules/CLI/PETStandardUptakeValueComputation/PETStandardUptakeValueComputation.cxx#L905
The above have confirmation in GE’s document, i.e.:
SUVlbm = (PET image Pixels) * (LBM in kg) * (1000 g/kg) / (injected dose)
…
tracer_activity = Total Dose (18,1074)
…
Total Dose(18,1074) = NET Activity to the patient at Series Time (0008, 0031)
To sum up, in order to calculate SUVmax for GE data, the following has to be calculated:
SUVlbm = [activity, i.e., signal for a single pixel]
* 1.10 * weight – 120 * (weight/height^2)
/
[tracer_activity * 2^( -(scan_time – measured_time)
/
half_life)]To be more precise, utilizing specific information about the voxel and DICOM metadata fields:
SUVlbm = [PET voxel (raw) data, e.g., from location: x=23, y=62, z=83]
* 1.10 * weight(from:0010,1010)
– 120 * (weight(from:0010,1010)/height(from:0010,1020)^2)
/
[dose(from:0018,1074)
* 2^( -(scan_time(from:0008,0031)
- measured_time(from:0018,1072))
/
half_time(from:0018,1075))]Note:
- `scan_time` in DICOM notation is `SeriesTime`, see: http://dicomlookup.com/lookup.asp?sw=Tnumber&q=(0008,0031)
- `measured_time` name in DICOM notation is `Radiopharmaceutical Start Time`, see: http://dicomlookup.com/lookup.asp?sw=Tnumber&q=(0018,1072)
- the discrepancy between GE’s documentation naming and variable naming in Slicer’s implementation. Generally Slicer is closer to the original DICOM nomenclature, cf., e.g., `startTimeSeconds` (Slicer) and `Radiopharmaceutical Start Time` (DICOM; see: http://dicomlookup.com/lookup.asp?sw=Tnumber&q=(0018,1072))
Please also note that the above formula is for males, for females, see information above.
April 17-18, Poznan, Poland; revised June 15, 2023
There is also a very important Slicer module extension: SUV Factor Calculator, with the source code available at: https://github.com/QIICR/Slicer-PETDICOMExtension/tree/master/SUVFactorCalculatorCLI.
Notice in particular the following file: https://github.com/QIICR/Slicer-PETDICOMExtension/blob/master/SUVFactorCalculatorCLI/SUVFactorCalculator.cxx, and the DecayCorrection function — https://github.com/QIICR/Slicer-PETDICOMExtension/blob/master/SUVFactorCalculatorCLI/SUVFactorCalculator.cxx#L173-L188. There is an important decayedDose part there:
double decayedDose = injectedDose * (double)pow(2.0, -(decayTime / halfLife) );which in GE’s document is called actual_activity.
The decayTime is calculated in the following way:
double decayTime = scanTimeSeconds - startTimeSeconds;In other words, it says how many seconds has passed since the beginning of the examination (injecting the marker) to the moment when the image was acquired.
In the example I analyze, there are the following constants (values/parameters):
INJECTED DOSE: 4.39e+08
RAD. START TIME: 37740
SERIES TIME: 41902
DECAYED DOSE: 283327(An output from the Slicer 3D’s SUV Factor Calculator module, as viewed after running the PET Standard Uptake Value Computation module on an ROI.)
Additionally, the startTimeSeconds is 37740.
In my example, the calculation is:
dose = 439000000
dose_in_kbq = dose * 0.001
actual_activity = dose_in_kbq * math.pow(2, (-((41902-37740)/6588)))
print(actual_activity)and the output is exactly the same as in Slicer, i.e.:
283327(rounded to the nearest decimal [int])
The lbm calculation is also quite straightforward, e.g..:
weight = 93
height = 164
leanBodyMass = 1.07*weight - 148*(weight/height)*(weight/height)Important note: height is in cm not in m.
Just to quickly sum up, in order to calculate SUVs (or SUVmax, if you are interested in a maximal value for a given ROI), you have to multiply the raw PET value (activity counts, that are in a scale of thousands) by a factor that is calculated – depending on the type of the factor – on all, or some of the following patient- and study-specific parameters: patient’s gender, height, weight, dose, half-time, injection time, and scan time (that will bring the values to the scale of dozens, as you would expect for SUVs).
The two most commonly used factors are: SUVbw and SUVlbm. Slicer’s PET Standard Uptake Value Computation module by default uses SUVbw, and, unfortunately doesn’t mention this in the interface. When we validated the results with outputs from the GE Work Station – which were using SUVlbm – we got inconsistencies in the results. Fortunately, Slicer has also the SUV Factor Calculator module, for which the output (i.e., the SUV Conversion Factors) is available after calculating the SUVmax for any ROI, see my video explaining how it works: https://youtu.be/JXpk3u9553I.
I wanted to convert the following activity count value into SUV: 145116.82. Importantly, because this value was the highest in the ROI I analyzed, it was in fact a SUVmax value for this ROI.
After taking into account the above mentioned patient- and study-specific parameters, I got the following SUV Conversion Factors: (1) SUVbw = 0.000310095; and (2) SUVlbm = 0.00022084.
Then the conversion is as simple as multiplying the activity count value by the conversion factor, i.e.:
SUVmax_bw = 145116.82 * 0.000310095 = 45.0000002979
SUVmax_lbm = 145116.82 * 0.00022084 = 32.0475985288Interestingly, the GE Work Station show the following SUVmax value for this ROI: 33.6 (about 5% mismatch) — we will be inquiring the issue further.
April 19, Poznan, Poland
One of the possible sources of discrepancy between the results is the fact that according to the GE PET/CT Discovery IQ Manual (page ~445) there are two possible SUVlbm coefficients: Legacy (Morgan et al., 1994 — pre-print of James et al., 1976; Sugawara et al., 1999) and Consensus (see James et al., 1976).
Legacy (Morgan et al., 1994; after: James et al., 1976):

The most probable source of the values for the “Consensus”, mentioned in the GE’s manual (only the value for male is stated there: 128) is the following quote form James et al., 1976:
cut-off points for obesity corresponding to a body fat of approximately 28 per cent for men and 34 per cent of total bodyweight for women
Consensus (based on James et al., 1976):
lbm_man = 1.10 * weight – 128 * (weight/height)^2
lbm_woman = 1.07 * weight – 134 * (weight/height)^2References
James WPT. Research on obesity. Her Majesty’s Stationery Office, London, 1976
Morgan DJ, Bray KM. Lean body mass as a predictor of drug dosage. Implications for drug therapy. Clin Pharmacokinet. 1994;26:292–307. doi: 10.2165/00003088-199426040-00005. [PubMed] [CrossRef] [Google Scholar]
Sugawara Y, Zasadny KR, Neuhoff AW, Wahl RL. Reevaluation of the standardized uptake value for FDG: variations with body weight and methods for correction. Radiology. 1999 Nov;213(2):521-5. doi: 10.1148/radiology.213.2.r99nv37521. PMID: 10551235.
April 23, Poznań, Poland
Interestingly, even in the Slicer source code, the authors were not sure which LBM factor is the proper one, see:
//leanBodyMass = 1.10*weight - 120*(weight/height)*(weight/height);
leanBodyMass = 1.10*weight - 128*(weight/height)*(weight/height); //TODO verify this formulaIn the Slicer implementation, there are the following LBM factors, per gender:
- Male:
128 - Female:
148
Unfortunately, according to my validation, performed on several patients, both male and female, the set of LBM factors that the most probably are implemented in GE scanners (work stations) is the following:
- Male:
120 - Female:
134
In other words, the optimal version seems to be a part-legacy, part-consensus (which probably Slicer was implementing at some point, see the commented line in the link above). The official GE manual states that the legacy version is used, but the 148 LBM factor value for woman seems to yield about 5% offset, in minus (the 134 works better). Further validation is needed. It is even possible to backward engineer the female (as well as the male) LBM factor, knowing GE results, patient metadata from the DICOM header, and the maximal value in the ROI (the raw, activity count values).
May 8, Poznan, Poland
I hope that you found this information useful, if yes, please follow me on LI for similar content: https://www.linkedin.com/in/mikolaj-buchwald/
Cover graphics: the photo by National Cancer Institute on Unsplash.